See our 2023 BioEssay for a view into some
of our recent research interests!
Host/Pathogen Interactions in the Intestine
How do organisms defend themselves against infection using ‘non-professional’ immune cells? In the Troemel lab we are addressing this question with the nematode host Caenorhabditis elegans. C. elegans provides a convenient system for studying intestinal infections, because it has a transparent body plan that enables visualization of infection in live animals. C. elegans also has a short lifespan and many genetic and molecular tools. Furthermore, C. elegans has intestinal epithelial cells with structural and functional similarity to human intestinal epithelial cells, so what we learn in worms may be relevant to human intestinal health.
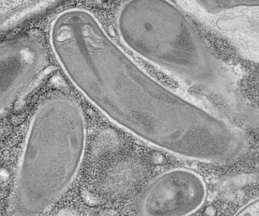
Microsporidia spores firing their polar tubes to deliver sporoplasms;
Troemel and Becnel, 2015
Current Interests
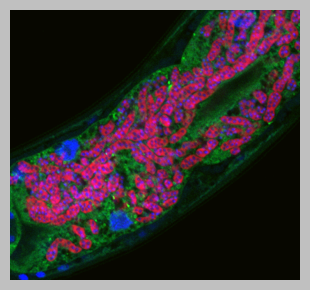
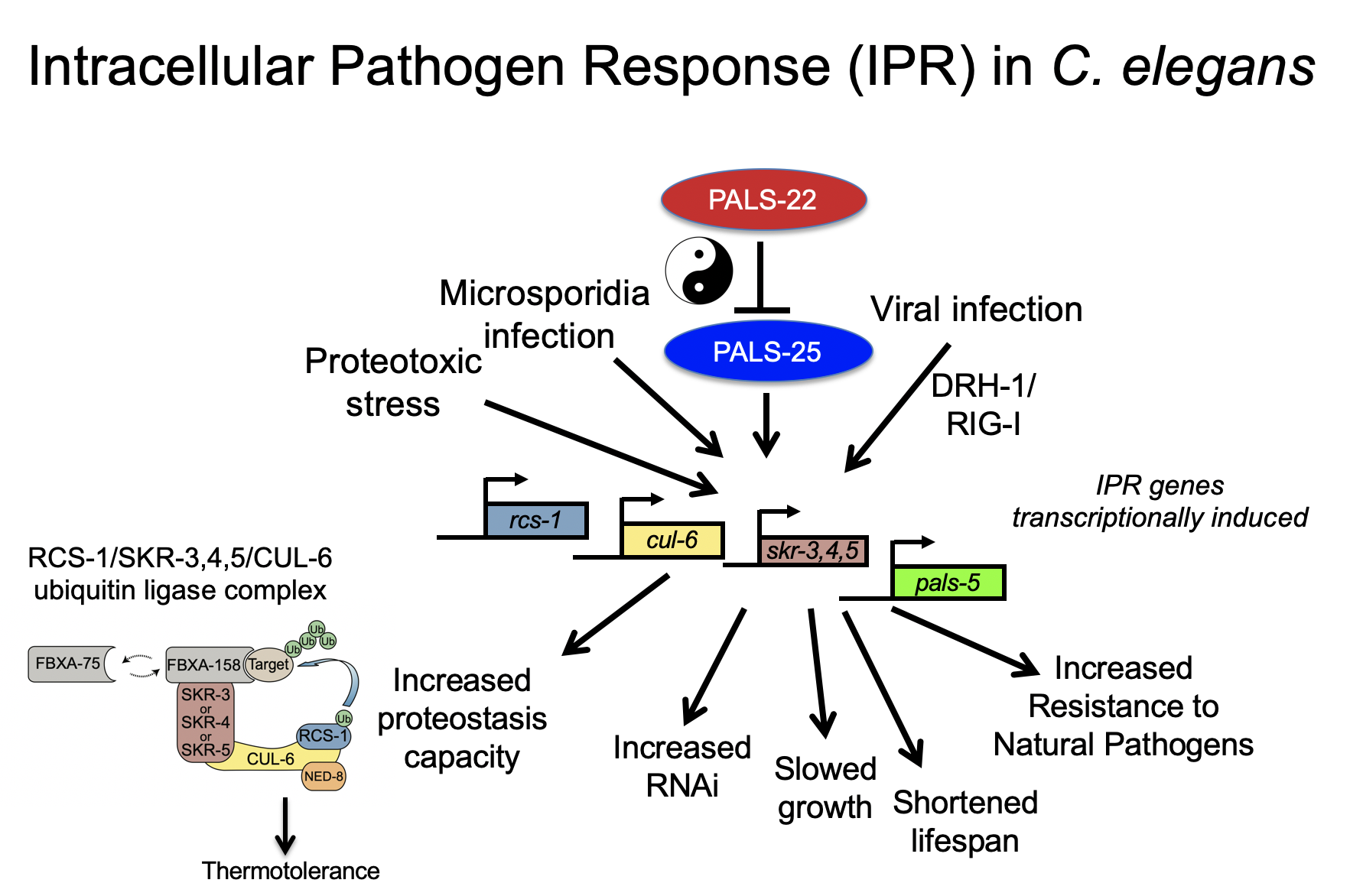
We have studied the C. elegans response to several different pathogens, but are currently focusing on response to microsporidia and the Orsay virus. Microsporidia comprise a phylum of fungal-related pathogens and are the most common known cause of infection for C. elegans in the wild. C. elegans responds to microsporidia infection with a robust transcriptional response. Surprisingly these microsporidia-induced genes are also induced by infection with the Orsay virus, which is a 3-gene RNA virus found infecting C. elegans in the wild (Bakowski et al, 2014). Microsporidia and the Orsay virus are molecularly distinct, but both are obligate intracellular pathogens of the intestine. We have named this common transcriptional response the “Intracellular Pathogen Response” and it appears to represent a physiological program that promotes defense at the expense of growth.
Current Projects
• RIG-I-like receptor DRH-1 senses RNA virus and activates the IPR
Relatively little is known about how C. elegans detects infection, and in fact many of the canonical pattern recognition receptors found in mammals are missing in C. elegans. One exception is the RIG-I-like receptors, which detect RNA viruses in mammals to induce transcription of type-I interferons. C. elegans has a RIG-I-like receptor called DRH-1, which previously had been shown by other groups to direct RNA interference. Our work has shown that DRH-1 also directs a transcriptional response, the IPR, upon detection of viral RNA replication products (Sowa et al, 2020). More recently, we have found that DRH-1 has previously unappreciated 2CARD signaling, which activates the IPR, similar to RIG-I 2CARD activation of the type-I interferon response (Batachari et al, 2024).
• Immunity/growth switch controlled by regulators like pals-22 and pals-25
We have studied the effects of IPR activation through analysis of mutants that have misregulated expression of IPR genes. For example, pals-22 mutants have upregulated IPR gene expression in the absence of infection (Reddy et al, 2017), and these mutants have dramatic changes in physiology, with increased resistance against microsporidia and virus infection, among other phenotypes (see below). Nearly all of the pals-22 phenotypes are reversed by mutations in the pals-25 gene, which is in an operon with pals-22. Thus wild-type pals-22 is a repressor and wild-type pals-25 is an activator of the IPR program, and we refer to them as ‘antagonistic paralogs’ (Reddy et al, 2019). We have recently found another pals module comprised of pals-17/pals-16/pals-20 (Lazetic et al, 2023), and also characterized a pals-25 gain-of-function mutant, which exhibits increased immunity including epidermal to intestinal signaling (Gang et al, 2022).
• Improved proteostasis mediated by a cullin-ring ubiquitin ligase
One striking phenotype in pals-22 mutants is increased tolerance of thermal stress and lowered levels of protein aggregates. Both of these phenotypes can be reversed by loss of one IPR gene, the cullin cul-6, which is predicted to act in a cullin ring ligase complex. We have found binding partners for the CUL-6 protein that act with CUL-6 to promote proteostasis capacity (Panek et al, 2020), and we have identified HSP-90 as a candidate substrate for the CUL-6 ligase (Bardan Sarmiento, 2024). We are continuing to define the components of this complex, as well as the targets they ubiquitylate, to understand how the IPR regulates protein homeostasis.
Other Areas of Research
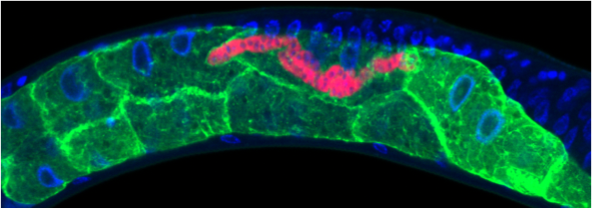
• Restructuring of C. elegans host cell biology by microsporidia infection
• Microsporidia transformation and nanoluciferase expression
• Microsporidia species identification, genomics and proteomics
• Natural variation in C. elegans resistance to Nematocida parasites
• C. elegans detects P. aeruginosa infection by sensing translational inhibition
P. aeruginosa is a Gram-negative bacterial pathogen that is a leading cause of death in cystic fibrosis patients and burn patients. Using an intestinal infection model in C. elegans, we have described how C. elegans detects translational inhibition caused by P. aeruginosa Exotoxin A, and uses the transcription factors ZIP-2 and CEBP-2 to mediate induction of defense gene expression (Dunbar et al, 2012; Reddy et al, 2016).